Frequently asked questions
Our team has assembled a list of frequently asked questions to help you find answers quickly. Filter using one or more categories to focus on specific topics, or use the search bar to perform a text search.
Search all FAQs:
What is the PAM sequence for CRISPR and where is it?
CRISPR-Cas9 mechanisms recognize DNA targets that are complementary to a short CRISPR RNA (crRNA) sequence. The part of the crRNA sequence that is complementary to the target sequence is known as a spacer. In order for Cas9 to function, it also requires a specific protospacer adjacent motif (PAM) that varies depending on the bacterial species of the Cas9 gene. The most commonly used Cas9 nuclease, derived from S. pyogenes, recognizes a PAM sequence of NGG that is found directly downstream of the target sequence in the genomic DNA, on the non-target strand.
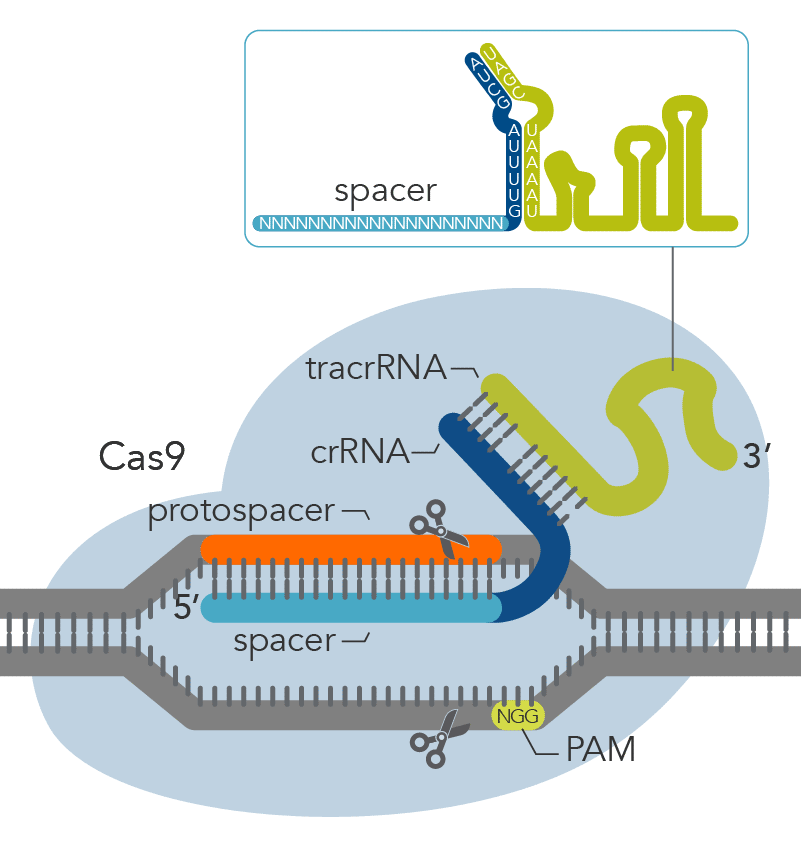
Recognition of the PAM by the Cas9 nuclease is thought to destabilize the adjacent sequence, allowing interrogation of the sequence by the crRNA, and resulting in RNA-DNA pairing when a matching sequence is present [1,2]. Cas9 nucleases with alternative PAMs have also been characterized and successfully used for genome editing [3]. It is important to note that the PAM is not present in the crRNA sequence but needs to be immediately downstream of the target site in the genomic DNA.
Want to Learn More?
Download The CRISPR Basics Handbook that covers applications of CRISPR technology from guide RNA design to data analysis.
The latest CRISPR abbreviations and definitions in a single list!
Design custom solutions for CRISPR genome editing.
References:
- Anders C, Niewoehner O, et al. (2014) Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature, 513(7519):569–573.
- Sternberg SH, Redding S, et al. (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature, 507(7490):62–67.
- Esvelt KM, Mali P, et al. (2013) Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nature Methods, 10(11):1116–1121.